Journal of Food Science and Nutrition Therapy
One health concept, prevalence and phenotypic antibiotic susceptibility of Escherichia coli and Salmonella isolated from meats sold in Lagos, Nigeria
Adebesin IO1*, Sule IO2, Kolapo KT3, Amoka SO2, Olomoko CR2 and Olubunmi OH1
2Department of Microbiology, Faculty of Life Sciences, University of Ilorin, Nigeria
3International Institute of Tropical Agriculture (IITA), Ibadan, Nigeria
Cite this as
Adebesin IO, Sule IO, Kolapo KT, Amoka SO, Olomoko CR, et al. (2023) One health concept, prevalence and phenotypic antibiotic susceptibility of Escherichia coli and Salmonella isolated from meats sold in Lagos, Nigeria. J Food Sci Nutr The 9(1): 024-033. DOI: 10.17352/jfsnt.000044Copyright
© 2023 Adebesin IO, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.This study reports the one health concept, prevalence, and antimicrobial susceptibility patterns of Salmonella and Escherichia coli isolated from raw and ready-to-eat (RTE) meats sold in cities of Lagos State, Nigeria. The conventional method of isolation was used to isolate E. coli and Salmonella spp. on their respective selective media from fifty meat samples obtained from abattoirs, open display, and packaged products at various locations in the state and was confirmed by Gram’s reaction and biochemical tests. Thirty-three E. coli and Twenty-seven Salmonella spp. were isolated with the overall prevalence rate recorded as 72% and 68% respectively. The isolated bacteria were subjected to antimicrobial susceptibility testing on nine different antibiotics using the agar disc diffusion method. All the Salmonella were resistant to at least one antibiotic while two E. coli isolates showed susceptibility to all the antibiotics used in this study. Of the 33 E. coli subjected to antimicrobial testing, 84.8% were susceptible to gentamicin, 81.8% susceptible to ciprofloxacin, and 75.8% susceptible to Augmentin. A lower susceptibility pattern was observed on Salmonella with 74.1% of the Salmonella being susceptible to ciprofloxacin and gentamicin, and 70.4% susceptible to azithromycin. Gentamicin was the most effective antibiotic while amoxicillin was found to be least effective against E. coli and Salmonella isolated from the meat samples used in this study. The multiple antibiotic resistance (MAR) index of Salmonella ranged between 0.11-0.67 while E. coli ranged between 0-0.89. E. coli was found to be more resistant than Salmonella and the bacteria isolated from RTE meats showed higher MAR than those isolated from raw meats.
Introduction
E. coli and Salmonella are Gram-negative bacteria belonging to the Enterobacteriaceae family that are commonly found in the intestinal tract of humans and animals [1]. These bacteria have been the causative agents of food poisoning, and urinary tract infections [2] and under severe conditions, they cause foodborne infections that could also lead to hemolytic colitis, hemolytic uremic syndrome, bacteremia, Reiter’s arthritis, and death [3]. The impact of these pathogens results in the loss of longer shelf life of food and huge monetary loss to the farmers and the sellers of products [4]. The negative impact of these diseases on human and animal populations cannot be overestimated as various outbreaks of E. coli and Salmonella have been reported in different countries of the world [5]. According to WHO [5], of the total foodborne illnesses that occur in the world, 40% affect children under 5 years old leading to 125,000 deaths per annum. Salmonella typhimurium was reported to be responsible for 115 million infections in humans and 370,000 deaths in the world per annum [6].
Infections by these bacteria are caused by the consumption of contaminated raw and poorly cooked food, feed in animals, and equipment such as utensils at homes and farm tools. Meats such as beef, pork, mutton, chicken, and their products have been reported to play major roles in the transmission of E. coli and Salmonella that are responsible for foodborne illnesses. Various studies have reported the presence of E. coli and Salmonella in different meats and their products [4,7,8]. Likewise, studies have shown that the level of E. coli and Salmonella prevalence in meats varies between regions as regions with better hygiene practices and proper handling of food and food products have lower prevalence and foodborne illness outbreaks.
Food poisoning infections require the use of antibiotics to alleviate the effects in both human and animal populations; however, bacteria have developed resistance to large numbers of antibiotics in use; thereby posing challenges that must be overcome. Also, the emergence of cross-resistance among pathogenic bacteria has greater consequences too. Cross-resistance which has been reported in many bacteria including E. coli and Salmonella is a process that occurs when resistance to one antibiotic causes resistance to another antibiotic of the same or different classes [9]. This cross-resistance is caused by different mechanisms such as genetic mutations, target site alteration, horizontal gene transfer, and efflux pumps [9]. This resistance and cross-resistance to antibiotics are attributed to the misuse, overuse, and excessive application of antibiotics in animals and humans; thereby leading the bacteria to modify means and mechanisms to overcome the constraint. This development of mechanisms to overcome antibiotic pressure has led to failure in the treatment of various bacterial-caused infections in humans and animals. To overcome the problem of food poisoning, the adoption of measures such as education on proper handling and processing, good agricultural practices, adherence to HACCP in the slaughtering and processing of meats for consumption, and proper disposal of waste should be adhered to [3]. Attention should be more focused on the prevention and control of food, feed, and equipment contamination by both food poisoning microbes and non-food poisoning microbes to maintain healthy living and retain the quality of food. The farmers, doctors, and drug administrators should not only be concerned about healing and curing diseases but also the effects of the overuse, misuse, and prolonged application on the environment should be considered.
The way the food production system, humans, animals, and the ecosystem/environment in which we live are interconnected is a prime example of the One Health Concept. Foods from animals play important roles in providing humans and animals with proteins and micronutrients. These animals have been reported to be responsible (hosts) for three out of five human diseases: zoonotic diseases [10]. Unhygienic environments have also been found to be breeding habitats for some animals that harbor diseases causing pathogens. Thereby, the concept illustrates that the health of people is connected to the health of animals, plants, and our shared environment. To promote the healthy existence of humans, the health of plants and animals that humans depend on for food and interact with, and the environment in which they are found must be maintained. Another layer of complexity to this complex issue is the role that the environment, especially soil and water, plays as a reservoir for bacteria resistant to antibiotics [11]. Antimicrobial resistance pathogens spread from animals to humans through these resistant strains’ ability to enter the human food chain. Since antimicrobial resistance must be addressed immediately, the One Health idea has been strengthened as infectious pathogens evade drugs meant to fight them. With the understanding that antimicrobial resistance is a complicated system woven across multiple fields, this holistic approach highlights the delicate relationship between the health of people, animals, plants, and the environment [12].
However, there is limited study and information available on the level of meat contamination and the spread of coliform, E. coli, and Salmonella among humans, animals, food, and environment in Lagos, Nigeria; whereby, there are loopholes in the handling before, during and after slaughtering of animals such as improper waste disposal, display over the counter, roadside selling, and inadequate heating were observed during this study. This study is based on the One Health viewpoint, the prevalence and phenotypic antibiotic susceptibility of Salmonella and Escherichia coli isolated from meats sold in Lagos, Nigeria. Through the evaluation of these bacteria’s prevalence and antibiotic resistance in meat sold in Lagos, the research contributes to the One Health goal of addressing antimicrobial resistance from a multidisciplinary perspective [13]. Given the growing impact of antimicrobial resistance, addressing this issue requires a unified front. Our goal is to add to the body of information required for creating strategies to slow the spread of antibiotic resistance across human, animal, and environmental interfaces by adopting the One Health approach and concentrating on the antimicrobial resistance landscape [14].
Materials and methods
Location of the study
This study was carried out in various cities in Lagos state, Nigeria. The state is estimated to have a land size of 3,577 square kilometers which culminates to 0.38% of Nigeria’s landmass. It shares boundaries with the Ogun state in the Northeast, the Bight of Benin in the south, and to the west the international border with the Republic of Benin.
Sampling of raw meats and Ready-to-Eat (RTE) meats for the study
In total, 50 meat samples were collected randomly and examined for the presence of coliform, E. coli, and Salmonella. These samples consisted of 5 each of raw chevon, raw guinea fowl, raw chicken, raw beef, raw mutton, RTE chevon, RTE guinea fowl, RTE chicken, RTE beef, and RTE mutton. Ten (10g) of each sample was collected in a sterile sampling bag (zip lock) and transported to the laboratory in a container containing ice packs. The samples were analyzed immediately in the laboratory.
Analysis of meat samples for coliforms, E. coli and Salmonella
Ten (10g) of the meat samples were pre-enriched in 90 ml peptone-buffered water and incubated at 37oC for 24 hours. 10ml of the homogenized culture was Pipetted into 90ml sterile distilled water to obtain 10-1 dilution. 10ml of 10-1 dilution was also transferred into another 90ml sterile distilled water to obtain 10-2 dilution. This procedure was repeated up to 10-4 dilution. For the culturing of raw meats aliquots, 0.1ml of the 10-2 to 10-4 aliquot was inoculated into MacConkey agar plate by spread plate technique for coliform count enumeration, and 0.1ml of 10-2 dilutions were streaked on Levine’s Eosin Methylene Blue Agar (EMB) and Xylose Lysine Deoxycholate agar (XLD) for E. coli and Salmonella detection respectively. For RTE meats, 0.1ml of the 10-1 to 10-3 aliquot was inoculated into Mac Conkey agar plate by spread plate technique and 0.1ml of 10-2 dilution was streaked on Levine’s Eosin Methylene Blue Agar and Xylose Lysine Deoxycholate agar. These plates were incubated at 37oC for 24 hours in an inverted position [15].
Confirmation of Escherichia coli and Salmonella Isolated from raw meats and RTE meats
The colonies that appeared dark centered and flat, with or without a green metallic sheen on Eosin Methylene Blue Agar were isolated and pre-summed to be E. coli. The presumed E. coli was purified on trypticase soy agar and incubated at 37°C for 24 hours. These isolates were confirmed using the Gram staining technique and biochemical tests such as Methyl red, Voges Paskeur, indole, citrate, urease, and hydrogen sulfide tests [15]. Also, the colonies that appeared red with or without a dark center were isolated on Xylose Lysine Deoxycholate agar and presumed to be Salmonella spp. These colonies were further confirmed using Salmonella-Shigella agar, Gram staining technique, Methyl red, Voges Paskeur, indole, citrate, urease, and hydrogen sulfide tests [15].
Phenotypic antibiotic susceptibility testing
Antibiotic susceptibility testing was carried out on the isolates using the disk diffusion method according to the Clinical and Laboratory Standards Institute [16]. Thirty-three E. coli and twenty-seven Salmonella were isolated and subjected to antibiotic testing using the following antibiotics: ciprofloxacin 5µg (CIP), chloramphenicol 30µg (C), tetracycline 30ug (T), ceftriaxone 30µg (CRO), gentamicin 10µg (GM), amoxicillin 30µg (A), Augmentin 30µg (AUG), azithromycin 15µg (ATH) and trimethoprim 2.5µg (TM). Culture suspension was made by inoculating 24 hours pure young colonies into 5ml of sterilized normal saline contained in a test tube. The turbidity of the normal saline was adjusted to 0.5 McFarland standard turbidity. The standardized inoculum was inoculated into already prepared Mueller Hinton Agar by swab stick to obtain dense and uniform growth. Sterile Forceps was used to introduce antibiotic discs onto the surface of the inoculated plates and the plates were incubated at 37oC for 24 hours. After incubation, the inhibition zones produced by antibiotics against the inoculum were measured in mm using a ruler and interpreted by CLSI [17].
Multiple Antibiotic Resistance (MAR) index
The MAR index of each Salmonella and E. coli isolate was determined by the ratio of the number of antibiotics that a particular bacterium was resistant (a) to the number of antibiotics tested (b). MAR was then calculated as a/b. The multidrug resistance profile was taken as being resistant to 3 or more antibiotics used in this study [18].
Results
Prevalence of coliforms, E. coli and Salmonella in meats sold in Lagos, Nigeria
The examination of the fifty samples collected aseptically showed to be positive for coliform (100%), 72% for E. coli, and 68% for Salmonella. The highest coliform count was reported in raw meat and the least in ready-to-eat meat. On average, raw meats had a higher coliform count than ready-to-eat meats. Raw beef had the highest count of 7.3×106 cfu/g and the lowest count on RTE chicken; 3×102cfu/g. The prevalence for E. coli and Salmonella ranged between 60-100% in all the meat samples. Guinea fowl was 100% positive for E. coli and Salmonella. In this study, 33 E. coli and 27 Salmonella were isolated from the samples positive for the respective bacteria. 22 and 11 E. coli were isolated from raw and RTE meats respectively and, 19 and 8 Salmonella from raw and RTE meats respectively Table 1.
Identification of the isolates by cultural methods
The bacteria were identified by the Gram staining method which showed that the isolates are Gram-negative. The biochemical test revealed E. coli to be positive for indole and methyl red test, negative for citrate, Voges Paskeur, hydrogen sulfide production, and urease tests. Salmonella showed positive results for methyl red and hydrogen sulfide tests, and negative for indole, citrate, Voges Paskeur, and urease tests Table 2.
Antimicrobial susceptibility pattern of E. coli isolated from meats sold in Lagos, Nigeria
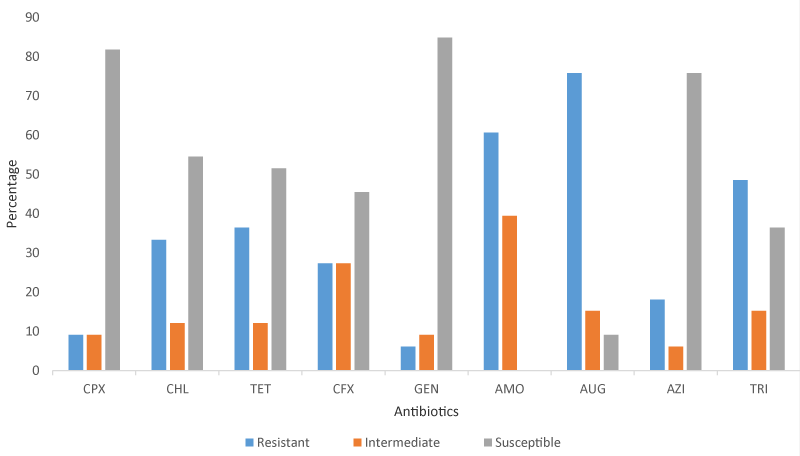
In this study, 33 E. coli isolated from both raw and RTE meats were subjected to an in-vitro antibiotic test against nine antibiotics. The least number of resistances to antibiotics by E. coli was observed on gentamicin while the highest was reported on amoxicillin and trimethoprim (12 each) in raw meat. In this study, none of the 33 E. coli isolated from both raw and RTE meats was susceptible to amoxicillin. Gentamicin, azithromycin, and ciprofloxacin are the three most effective antibiotics that the E. coli isolates were highly susceptible to in this study. Two E. coli isolated from RTE mutton and raw beef showed resistance to all the antibiotics used in this study. Despite gentamicin being one of the most potent, none of the E. coli isolated from ready-to-eat meats was susceptible to its antibacterial action Table 3, Figure 1.
Antimicrobial susceptibility pattern of Salmonella isolated from meats sold in Lagos, Nigeria
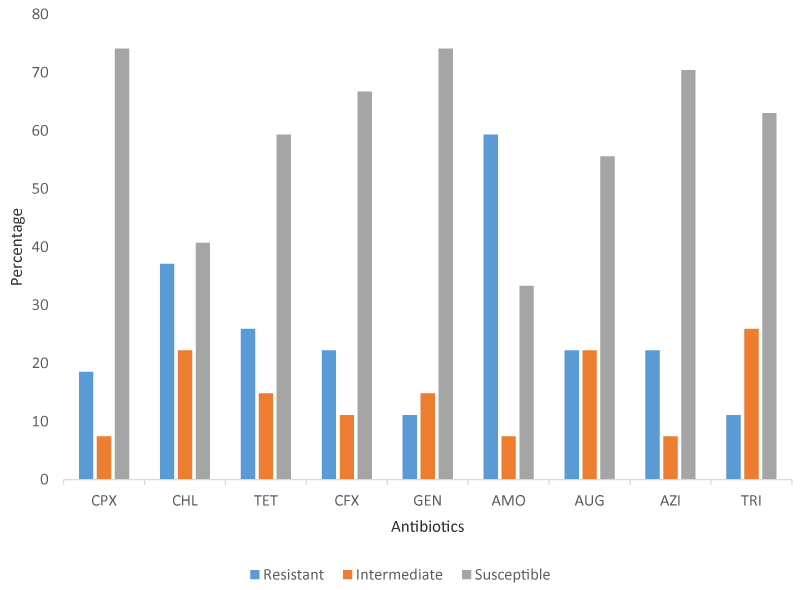
Among the 27 Salmonella used for the antibiotic test, nineteen Salmonella were isolated from raw meats and eight from RTE meats. Unlike E. coli, all the Salmonella were susceptible to at least one of the antibiotics used in this study. In raw meat, the highest resistance was recorded on amoxicillin (13/19) followed by chloramphenicol (8/19), and the least resistance on gentamicin (1/19). In RTE meats, 4 of the 8 Salmonella were resistant to tetracycline and augmentin while 1 was resistant to trimethoprim. Unlike E. coli isolated from raw meats, Salmonella isolated from RTE meats showed high susceptibility to amoxicillin Table 4.
Phenotypic Antibiotic susceptibility profile of Salmonella isolated from meats sold in Lagos, Nigeria (Figure 2)
Multiple Antibiotic Resistance index (MAR) index of E. coli
The MAR of E. coli ranged between 0 to 0.89. Two E. coli isolated from beef and RTE mutton were susceptible to all the antibiotics used. 9 out of the 33 E. coli were resistant to 3 antibiotics which is the most frequent MAR recorded. The highest MAR recorded is 0.89 by E. coli isolated from RTE chicken Table 5.
Multiple Antibiotic Resistance index (MAR) index of Salmonella
The MAR index recorded in this study for Salmonella ranged between 0.11 to 0.67. The highest percentage (11/27) of Salmonella subjected to the antibiotic test exhibited resistance to two antibiotics. One isolate each showed resistance to 4, 5, and 6 of the antibiotics. Six Salmonella showed resistance to 1 antibiotic while seven were resistant to three antibiotics used in this study Table 6.
Discussion
The meats were expected to be free of bacteria and the highest population of coliform was recorded in beef. All the meats used in this study were positive for coliform contamination which is close to the value reported by Maharjan, et al. [19] who concluded that more than 80% of the meats collected were positive for coliform contamination. Coliform contamination of 3.52 × 107 cfu/g for chevon and 2.14 × 107 cfu/g for beef was reported by Antwi-Agyei and Maalekuu [20] which were higher than the level reported in this study. A lower coliform count of 3.12 logcfu/ml was recorded by Sowunmi, et al. in a study on raw meats obtained from Sabo Market in Ikorodu, Lagos State [21]. Kim and Yim reported an average coliform count of 0.37logcfu/g which is lower than the range documented in this study on the raw and RTE meats [22].
The phenotypic appearance of E. coli and Salmonella colonies on their respective selective media, Gram’s reaction, and cultural biochemical tests profile were similar to the features reported by Feng, et al. [15]; and Raji, et al. [23]. These pathogens are known to reside in the gastrointestinal tract of humans and other animals, live close to one another, and circulate throughout the ecosystem through defecation, death of the harboring animal, and improper disposal of the waste. Bacteria in this close association tend to share and transfer traits like resistant genes among one another [24]. These pathogens are also known to get to the environment during slaughtering and get transferred to humans and other animals through contaminated feed, food, water, utensils, and uncooked and improperly cooked meats. The contamination of the meat and utensils used during the slaughtering of animals and animal processing has led to the transfer of these pathogens to humans and caused various enteric diseases that sometimes result in the death of humans in the absence of adequate medical attention. The consumption of meats and meat products contaminated with bacterial foodborne pathogens can lead to foodborne infections and outbreaks which are of great concern to the public health and public health experts. The contamination of these products with bacteria will result in short shelf life or early spoilage and unavoidable financial loss to farmers, sellers, and consumers [4].
Escherichia coli and Salmonella are some of the most important bacterial pathogens of humans and animals that can be transferred to meats and meat products and cause various foodborne infections, sickness, and death. The detection of E. coli and Salmonella in meat samples used in this study indicates the presence of lapses in the process of slaughtering the animals, handling, transportation, cooking, and display at markets [25]. The high prevalence of these bacteria could also be attributed to a lack of proper hygiene during the slaughtering of the animal as there are possibilities of the intestinal content ruptures to come in contact with the meats, surfaces, and utensils used during evisceration. Also observed during sampling was the proximity location of the waste dumping site to the preparation point contributing to the contamination as flies were seen settling on the meats. Another possible means of cross-contamination observed during sampling was the knives used for evisceration were also used to cut the meat into pieces. In the market, the meats are always on display in the open air without covering which brings potential pathogen-laden houseflies in contact with the meats. During the purchase of meats (beef, mutton) in the market, it is a common practice of potential buyers to inspect many meat portions with bare hands before a choice is made. Among the samples is a local meat preparation sold in Nigeria as ‘suya’. The analysis of the sample yielded the least coliform population among all the samples analyzed despite the meat being exposed to naked fire. Analysis of the suya yielded no E. coli and Salmonella growth in this study.
In this study, E. coli was absent in 14 (28%) meats while Salmonella was absent in 16 meats (32%). This high prevalence clearly shows the danger associated with meat consumption. A similar level of E. coli contamination in retail chicken (83.5%) and beef (68.9%) was reported by Zhao, et al. [26] in the US. Kim and Yim [22] conducted a study on various livestock meats sold in the Korean Republic, E. coli was found to be absent, but coliforms were detected. The absence of these pathogens is the reflection of adherence to the practices suggested by Adzitey at the time of the study [27]. The situation is subject to changes as there is a possibility of opposite results in future studies that may result from management changes, non-adherence to the quality management system, and poor hygiene. A lower value of E. coli contamination was reported by Adzitey [27]; and Altalhi, et al. [28] on various meat samples studied in comparison with this study. In a study conducted by EFSA [29] on meat products and meat preparations from mixed sources, low levels of contamination of 2.7% by non-0157 E. coli and 2.2% by Shiga toxin-producing E. coli were reported. Ciekure, et al. [30] studied ready-to-eat meat (RTE) collected in Lavia, it was discovered that 32% of the meat products were contaminated with Escherichia coli despite being exposed to heat. Also, 54% level of E. coli contamination was detected in raw beef in a study by Adzitey [27] and 76% in a study by Parvin, et al. [31] on frozen chicken meat. Adeyanju and Ishola [32] obtained a lower prevalence of 33% for Salmonella and 43.4% for Escherichia coli isolated from retailed poultry meat and processed meats sold in Oyo State, Nigeria. In a comparative study to determine the level of E. coli contamination between locally made chicken meat and chicken thigh imported into Ghana, the study showed higher levels of E. coli contamination on local chicken (64.29%) than on imported chicken (55.30%) Rasmussen, et al. [33]. Another study on the prevalence of E. coli O157:H7 from beef in Kwara State Nigeria, showed a 5.6% prevalence which is lower than the 80% recorded in this study [34]. A higher prevalence (56%) of E. coli than Salmonella (31%) was reported by Adzitey, et al. which agrees with our study’s result [35]. A similar study on E. coli obtained from meats in Sabo, Ikorodu region of Lagos state showed 82% overall prevalence (higher than this study prevalence by 10%) with raw sheep and raw Guinea fowl (87.5%) each, raw beef (85%), raw local chicken (77.5%) and raw goat (72.5%) [21].
In this study, a higher percentage of contamination was discovered in raw meats than in ready-to-eat meats (RTE). The lower percentage of these bacteria in RTE is attributed to the process of cooking, frying, and drying that exposed the meat to high temperatures and killed the bacteria present in the meat together. It is expected that the level of contamination of meat in countries where proper adherence to handling and processing protocol of animals into meat to be lower than in developing countries with low regard to proper handling and hygiene practices. The process of preventing and controlling bacterial foodborne pathogens begins with proper handling and hygiene from slaughtering houses to the last link of consumption at various homes and offices. Minimizing the presence of bacteria including coliforms, fecal coliforms, Salmonella, and other pathogenic bacteria is vital in ensuring the quality health of the populace. The control and preventive measures described by Adzitey [36] must be adhered to in order to prevent foodborne infections. Good agricultural practices during activities such as animal breeding, rearing, slaughtering, transporting animals, etc. are the first line of action to prevent foodborne diseases caused by bacteria Adzitey [37]. Regular and proper waste disposal, and avoidance of contact between waste, equipment, food, and water have also been in practice to avoid contamination. Great emphasis should be placed on thorough hygienic handling and processing of animals into meat and meat products to reduce the level of morbidity and mortality caused by E. coli, Salmonella, and other pathogens.
The high level of resistance to commercial antibiotics in this study is a great concern as these bacteria have acquired different mechanisms to overcome the antibacterial effect. This high level of resistance recorded in this study and past studies revealed the high level of risks associated with the overuse and the abuse of antibiotics in animal husbandry and these bacteria eventually make their way into the environment and human population and continue circulation between animals and humans. The consumption of these meats poses a great danger as the antibiotic-resistant bacteria are transferred to humans. It also leads to the accumulation of antibiotic residue and the spread of these antibiotic-resistant genes within the hosts which will result in drug failure when treating diseases caused by these pathogens in the future. In this study, only two E. coli isolated from beef and RTE mutton were susceptible to all the antibiotics used without any Salmonella isolated from RTE meat being susceptible to azithromycin. Gentamicin was the most potent antibiotic as it showed susceptibility of 84.8 and 74.1% against E. coli and Salmonella respectively. Zhao, et al. [26] reported higher levels of resistance (18.6%) by E. coli isolated from different retail meats in the USA to gentamicin. A higher susceptibility to ciprofloxacin (95.56%) and trimethoprim (82.22%) but a lower susceptibility to gentamicin (75.56%) was reported on E. coli isolated from raw meats in Ghana [38]. Adzitey, et al. [39] found the highest level of resistant E. coli in the hands of meat sellers which is similar to the high MAR index reported in the RTE meat reported in this study. In a study by Hassan, et al. [40], a total of 34 bacteria were isolated from raw chicken of which 97%, 94%, 64%, 50%, 32%, 24%, 21%, and 6% were resistant to oxacillin, penicillin, ampicillin, cefotaxime, tetracycline, erythromycin, ciprofloxacin, and gentamicin respectively. E. coli and Salmonella were also identified as one of the most prevalent bacteria with the highest resistance. Also, a study conducted on meat and its products in Kalobia, Egypt by Abd-El-Tawab, et al. [41] showed that 11 (6.3%) of the bacteria isolated were E. coli and were highly resistant to methicillin and oxacillin but sensitive to enrofloxacin and gentamicin. The finding of this study on the sensitivity of E. coli to gentamicin is similar to the result obtained by Hassan, et al. [40] and this present study. In a similar study by Sowunmi, et al. in Lagos State the MAR of E. coli isolated from raw meats ranged between 0-88 which agrees with MAR recorded on E. coli in this study [21]. Also, most of the E. coli from the study had MAR of 0.38 which is also close to 0.33 recorded for most E. coli in our study. Higher susceptibility values were recorded for gentamicin (88.3%), trimethoprim (85%), chloramphenicol (83.3%), and ceftriaxone (80%) were reported [21]. In another study by Adetunji and Ishola [42], the virulent strains of Salmonella and E. coli were found to have 60% resistance each while the non-virulent strains had a lower resistance of 50% each. Also, a study by Adzitey, et al. [43] in which 84.0, 88.9, 88.0, 89.0. 86.7, 80.0 and 75.6% prevalence of E. coli was reported in meat, mutton, guinea fowl, beef, local chicken, and chevon respectively with 85, 73.33 and 71.69% of E. coli being resistant to erythromycin, tetracycline, and ampicillin respectively while 68.33% (41/60) of the identified E. coli exhibited resistance to two or more antibiotics. Pławinska-Czarnak, et al. studied Salmonella obtained from raw meats in Poland and obtained 53.84% of several Salmonella strains to be multidrug resistant [44]. The MAR index of the study on Salmonella ranged between 0.09 -0.51 which falls in the MAR range recorded in our study [44]. A contrary report on E. coli was reported by Saud, et al. that tetracycline had resistance values of 60.6% while the resistance value reported for gentamicin agrees with this study result [45]. Another contrary result on E. coli isolated from raw meats showed 75% resistance to tetracycline [46].
The resistance to the 3rd generation cephalosporins; ceftriaxone by E. coli and Salmonella in this study poses a serious concern due to the extensive use of ceftriaxone in the treatment of diarrhea and Salmonellosis in humans. A similar report on Salmonella isolated from raw meats to be resistant to the third-generation cephalosporins family was documented by Plawinska-Czarnak, et al. [44]. This spread of resistance to 3rd generation cephalosporins and beta-lactam groups which are in high use in human medicine is worrying. AmpC and extended-spectrum beta-lactamase are usually antibiotic-resistant mediated genes in E. coli and Salmonella. 36.4% and 25.9% of E. coli and Salmonella were resistant to tetracycline despite the embargo imposed on its non-therapeutic use in animal treatment. Augmentin: an antibiotic that contains amoxicillin (beta-lactam family) and clavulanic acid (beta-lactamase inhibitor) showed greater antibacterial activity on both E. coli and Salmonella than amoxicillin. This higher activity can be attributed to the presence of clavulanic acid in Augmentin which created synergistic antibacterial action with amoxicillin on these pathogens. The high level of E. coli and Salmonella resistance to amoxicillin may be attributed to being the commonest and cheapest antibiotics in Nigeria’s locality which makes its use on a higher level than other antibiotics. It is widely used for the treatment of diarrhea in animal husbandry and human medicine, and this could have led to the acquisition and development of survival strategies.
The MAR index of E. coli ranged from 0-0.89 antibiotics while Salmonella ranged from 0-0.69 antibiotics. 2 E. coli isolates were found susceptible while none was found resistant to all the antibiotics used in this study. 4, 7, 9, and 3 E. coli showed MAR of 11%, 22%, 33%, and 44% while 4, 2, 1, and 1 E. coli had MAR indexes of 56%, 67%, 78%, and 89% respectively. None of the Salmonella was found susceptible and resistant to all the antibiotics used in this study. 6, 11, 7, 1, 1, and 1 Salmonella had MAR of 11%, 22%, 33%, 44%, 56%, and 67% respectively. Kathleen, et al. [18] reported that bacteria with a MAR index above 2 were from an environment with high antibiotic usage and bacteria with a MAR below 2 were from an environment with low utilization of antibiotics for disease treatment and prevention. The result of the antibiotic susceptibility of E. coli and Salmonella obtained from meats in this study can be categorized as multidrug-resistant as a high percentage of the bacteria exhibited resistance to more than two antibiotics. In this study, 61% of the E. coli had a MAR index between 0.33-0.88 which indicates that these bacteria are from environments where antibiotics are largely used, misused, and abused. A lower trend was observed in Salmonella of which 37% had a MAR range of 0.33-0.67. E. coli and Salmonella isolated from RTE meats possessed a higher MAR index with the highest source of contamination being attributed to human contact after cooking, frying, smoking, oven drying, etc. Humans are thereby harboring many of these multi-drug resistant Salmonella and E. coli which could cause failure in treating Salmonellosis and E. coli infections in the future. Adzitey, et al. [47] reported 41 (68.33%) of the E. coli isolated from livestock sold in Ghana metropolis to be multi-drug resistant to three or more antibiotics. Saud, et al. [45-36] reported 52.5% multi-drug resistance by E. coli isolated from meats and 69.81% multi-drug resistance by E. coli isolated from various samples used in the study. Altalhi, et al. [28] found that 86.5% of E. coli were resistant to at least one antibiotic and 40.5% were resistant to at least three antimicrobials. 68.33% MDR was reported for E. coli which is closer to the value reported in this study (61%) [21]. Slaughtering of animals only after the withdrawal days of antibiotic treatment is a way out as it will reduce the spread of antibiotic residue in the animals to humans and antibiotic-resistant bacteria. Ekli, et al. reported that 73.2% of farmers in Wa municipality of Ghana did not practice the withdrawal method before animals were slaughtered for consumption [48]. The consumption of animals killed before the withdrawal day of antibiotic treatment will lead to the accumulation of these antimicrobial residues in the body which the intestinal flora will become adapted to. This will possibly lead to later time failure of such antibiotics in disease treatment.
The data obtained in this study poses serious challenges to one health concept as E. coli and Salmonella were found to be present in meats meant for public consumption. These bacteria were also found to be multidrug resistant. The antibiotic-resistant genes contained in E. coli and Salmonella can be transmitted to the gut microbiota of humans and create more antibiotic-resistant strains in humans. These antibiotic-resistant bacteria will later be released into the environment through defecation which will lead to the continuous spread of antibiotic-resistant bacteria and the exchange of antibiotic-resistant genes in the environment. This cycle continues again through the ingestion of contaminated forage, water, and soil by animals which are then consumed by humans and then to the environment. The health of the environment is critical in maintaining the health and well-being of plants, humans, and animals. Unsafe water, poor sanitation, and poor hygiene are responsible for human and animal mortality and morbidity, particularly in vulnerable populations in low-income countries.
Nonetheless, these results must be interpreted with caution and limitations should be borne in mind. The findings of this study are time-based as results from future studies may show deviation due to hygiene practices and adherence to the withdrawal method. Also, this study does not give the trueness of the E. coli and Salmonella prevalence in Lagos state as few cities were considered for the study, hence, future studies are encouraged to focus on the state at large and higher number of samples. In addition, there is little or lack of data available or prior research on similar studies in the state, thereby the need for future research to give more insight into these pathogens’ prevalence and susceptibility to antibiotics. The virulence factors and genes responsible for the resistance in E. coli and Salmonella were not investigated due to the limitations of the available facilities in the laboratory. Further studies are encouraged for the determination of genes responsible for the resistance to these antibiotics, the mechanisms used to transfer these genes, and the possibility of drug combination as seen in Augmentin. Also, future studies should be directed towards determining the possibility of cross-resistance between these Salmonella and E. coli within the host.
Conclusion
Overall, 50 (100%) of the meat samples were positive for coliform with 72 and 68% positive for E. coli and Salmonella respectively. Raw meats were contaminated more than RTE meats which shows a reduction in the contamination level due to the exposure to heat during processing. However, adequate hygiene practices are recommended for RTE meat sellers to prevent the spread of pathogenic bacteria among humans, animals, and the environment. The phenotypic antibiotic resistance revealed E. coli to be highly resistant to Augmentin, amoxicillin, and trimethoprim, and high susceptibility to ciprofloxacin, gentamicin, and azithromycin. Also, Salmonella showed high levels of resistance to amoxicillin and high susceptibility to all the antibiotics except amoxicillin and mild to chloramphenicol. To overcome the problem of high resistance to antibiotics observed in this study, there is a need for farmers to practice the withdrawal method, reduce the level of antibiotic use, and practice proper hygiene and management practices. This is to reduce the use of antibiotics in treating diseases and the spread of antibiotic-resistant bacteria among animals and the environment. An organic system of preventing and treating diseases should be adopted among the farmers. It is also recommended that farmers, butchers, meat sellers, and the public be given proper awareness regarding good hygiene practices. The government authority from the local to the federal level should establish a body that will exercise regular meat inspection before killing and distribution to the public.
- Center for Control and Disease Prevention (CDC) (2014). E. coli (Escherichia coli). https://www.cdc.gov/ecoli/general/index.html (accessed on 23 August 2020).
- Center for Control and Disease Prevention (CDC) (2020). Outbreak of E. coli infections. https://www.cdc.gov/ecoli/2020/o157h7-10-20a/index.html (accessed on 23 August 2020).
- Adams R, Moss MO. Food microbiology. RSC Publishing. Cambridge, UK. 2008; 1-436.
- Adzitey F, Teye GA, Kutah WN, Adday S. Microbial quality of beef sold on selected markets in the Tamale Metropolis in the northern region of Ghana. Livestock Res Rural Dev. 2011; 23:8-13.
- World Health Organization. Outbreaks of E. coli O104:H4 Infection. (2020). https://www.euro.who.int/en/health-topics/disease-prevention/food-safety/outbreaks-of-e.-coli-o104h4-infection (accessed on 23 August 2020).
- Seif Y, Kavvas E, Lachance JC, Yurkovich JT, Nuccio SP, Fang X, Catoiu E, Raffatellu M, Palsson BO, Monk JM. Genome-scale metabolic reconstructions of multiple Salmonella strains reveal serovar-specific metabolic traits. Nat Commun. 2018 Sep 14;9(1):3771. doi: 10.1038/s41467-018-06112-5. PMID: 30218022; PMCID: PMC6138749.
- Waters AE, Contente-Cuomo T, Buchhagen J, Liu CM, Watson L, Pearce K, Foster JT, Bowers J, Driebe EM, Engelthaler DM, Keim PS, Price LB. Multidrug-Resistant Staphylococcus aureus in US Meat and Poultry. Clin Infect Dis. 2011 May;52(10):1227-30. doi: 10.1093/cid/cir181. Epub 2011 Apr 15. PMID: 21498385; PMCID: PMC3079400.
- Bostan K, Aydin A, Ang MK. Prevalence and antibiotic susceptibility of thermophilic Campylobacter species on beef, mutton, and chicken carcasses in Istanbul, Turkey. Microb Drug Resist. 2009 Jun;15(2):143-9. doi: 10.1089/mdr.2009.0894. PMID: 19432522.
- Sincak M, Šoltisová K, Luptakova A, Sedlakova-Kadukova J. Overproduction of efflux pumps as a mechanism of metal and antibiotic cross-resistance in the natural environment. Sustainability. 2023; 15: 8767.
- World Organization for Animal Health. (2019) One Health “at a Glance”. http://www.oie.int/en/ for-the-media/onehealth. Retrieved on 22-02-2020.
- EFSA Panel on Biological Hazards (BIOHAZ); Koutsoumanis K, Allende A, Álvarez-Ordóñez A, Bolton D, Bover-Cid S, Chemaly M, Davies R, De Cesare A, Herman L, Hilbert F, Lindqvist R, Nauta M, Ru G, Simmons M, Skandamis P, Suffredini E, Argüello-Rodríguez H, Dohmen W, Magistrali CF, Padalino B, Tenhagen BA, Threlfall J, García-Fierro R, Guerra B, Liébana E, Stella P, Peixe L. Transmission of antimicrobial resistance (AMR) during animal transport. EFSA J. 2022 Oct 25;20(10):e07586. doi: 10.2903/j.efsa.2022.7586. PMID: 36304831; PMCID: PMC9593722.
- Fitzpatrick KJ, Rohlf HJ, Sutherland TD, Koo KM, Beckett S, Okelo WO, Keyburn AL, Morgan BS, Drigo B, Trau M, Donner E, Djordjevic SP, De Barro PJ. Progressing Antimicrobial Resistance Sensing Technologies across Human, Animal, and Environmental Health Domains. ACS Sens. 2021 Dec 24;6(12):4283-4296. doi: 10.1021/acssensors.1c01973. Epub 2021 Dec 7. PMID: 34874700.
- Oloso NO, Fagbo S, Garbati M, Olonitola SO, Awosanya EJ, Aworh MK, Adamu H, Odetokun IA, Fasina FO. Antimicrobial Resistance in Food Animals and the Environment in Nigeria: A Review. Int J Environ Res Public Health. 2018 Jun 17;15(6):1284. doi: 10.3390/ijerph15061284. PMID: 29914203; PMCID: PMC6025306.
- Falkenberg T. Applying a One Health approach to inter- and trans-disciplinary research on antimicrobial resistance. European Journal of Public Health. 2019; 29. 10.1093/eurpub/ckz185.798.
- Feng P, Weagant SD, Jinneman K. Bacteriological Analytical Manuel Chapter 4A: Diarrheagenic Escherichia coli. 2019. https://www.fda.gov/food/laboratory-methods-food/bam-chapter-4a-diarrheagenic-escherichia-coli.
- Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. 29th edition. CLSI Supplement M100. Wayne, Pennsylvania, US: Clinical and Laboratory Standards Institute. 2019; 26.
- Clinical and Laboratory Standards Institute. Clinical Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. 30th ed. CLSI supplement M 100, Wayne, PA, USA. 2020.
- Kathleen MM, Samuel L, Felecia C, Reagan EL, Kasing A, Lesley M, Toh SC. Antibiotic Resistance of Diverse Bacteria from Aquaculture in Borneo. Int J Microbiol. 2016;2016:2164761. doi: 10.1155/2016/2164761. Epub 2016 Sep 25. PMID: 27746817; PMCID: PMC5055980.
- Maharjan M, Joshi V, Joshi DD, Manandhar P. Prevalence of Salmonella species in various raw meat samples of a local market in Kathmandu. Ann N Y Acad Sci. 2006 Oct;1081:249-56. doi: 10.1196/annals.1373.031. PMID: 17135520.
- Antwi-Agyei P, Maalekuu BK. Determination of microbial contamination in meat and fish products sold in the Kumasi metropolis (a case study of Kumasi central market and the Bantama market). Merit Research Journal of Agricultural Science and Soil Sciences. 2014; 2:38–46.
- Sowunmi K, Olamiji OM, Adesola DY, Lawal AA, Adejoke OK. Antimicrobial Resistance of Escherichia coli Isolated from various Meat types in Sabo Market Ikorodu, Lagos. J Appl Microb Res. 2022; 5(1):14-19.
- Kim JH, Yim DG. Assessment of the Microbial Level for Livestock Products in Retail Meat Shops Implementing HACCP System. Korean J Food Sci Anim Resour. 2016 Oct 31;36(5):594-600. doi: 10.5851/kosfa.2016.36.5.594. PMID: 27857534; PMCID: PMC5112421.
- Raji M, Adekeye J, Kwaga J, Bale J, Heton M. Serovar and biochemical characterization of E. coli isolated from colibacillosis cases and dead in shell embryo in poultry in Zaria, Nigeria. Veterinarski Archiv. 2007; 77(6): 495-505.
- Peterson E, Kaur P. Antibiotic Resistance Mechanisms in Bacteria: Relationships Between Resistance Determinants of Antibiotic Producers, Environmental Bacteria, and Clinical Pathogens. Front Microbiol. 2018 Nov 30;9:2928. doi: 10.3389/fmicb.2018.02928. PMID: 30555448; PMCID: PMC6283892.
- Ritchie H, Roser M. Meat and dairy production 2019. https://ourworldindata.org/meat-production#number-of_animals-slaughtered.
- Zhao S, Blickenstaff K, Bodeis-Jones S, Gaines SA, Tong E, McDermott PF. Comparison of the prevalences and antimicrobial resistances of Escherichia coli isolates from different retail meats in the United States, 2002 to 2008. Appl Environ Microbiol. 2012 Mar;78(6):1701-7. doi: 10.1128/AEM.07522-11. Epub 2012 Jan 13. PMID: 22247155; PMCID: PMC3298142.
- Adzitey F. Prevalence of Escherichia coli and Salmonella spp. in beef samples sold at Tamale Metropolis, Ghana. International Journal of Meat Science. 2015; 20(5): 8–13.
- Altalhi AD, Gherbawy YA, Hassan SA. Antibiotic resistance in Escherichia coli isolated from retail raw chicken meat in Taif, Saudi Arabia. Foodborne Pathog Dis. 2010 Mar;7(3):281-5. doi: 10.1089/fpd.2009.0365. PMID: 19911929.
- European Food Safety Authority and European Centre for Disease Prevention and Control (EFSA and ECDC). The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2017. EFSA J. 2018 Dec 12;16(12):e05500. doi: 10.2903/j.efsa.2018.5500. PMID: 32625785; PMCID: PMC7009540.
- Ciekure E, Siksna I, Valcina O, Viksna L, Krumina A. Microbiological quality of ready-to-eat products and potential risks for consumers in Latvia. Nat Exact Appl Sci. 2016; 70: 245–251.
- Parvin MS, Talukder S, Ali MY, Chowdhury EH, Rahman MT, Islam MT. Antimicrobial Resistance Pattern of Escherichia coli Isolated from Frozen Chicken Meat in Bangladesh. Pathogens. 2020 May 28;9(6):420. doi: 10.3390/pathogens9060420. PMID: 32481680; PMCID: PMC7350304.
- Adeyanju GT, Ishola O. Salmonella and Escherichia coli contamination of poultry meat from a processing plant and retail markets in Ibadan, Oyo State, Nigeria. Springerplus. 2014 Mar 12;3:139. doi: 10.1186/2193-1801-3-139. PMID: 25674440; PMCID: PMC4320193.
- Rasmussen MM, Opintan JA, Frimodt-Møller N, Styrishave B. Beta-Lactamase Producing Escherichia coli Isolates in Imported and Locally Produced Chicken Meat from Ghana. PLoS One. 2015 Oct 13;10(10):e0139706. doi: 10.1371/journal.pone.0139706. PMID: 26461270; PMCID: PMC4603670.
- Ajuwon BI, Babatunde SK, Kolawole OM, Ajiboye AE, Lawal AH. Prevalence and antibiotic resistance of Escherichia coli O157:H7 in beef at a commercial slaughterhouse in Moro, Kwara State, Nigeria. Access Microbiol. 2021 Nov 11;3(11):000289. doi: 10.1099/acmi.0.000289. PMID: 35018331; PMCID: PMC8742587.
- Adzitey F, Teye GA, Anachinaba IA. Microbial quality of fresh and smoked guinea fowl meat sold in the Bolgatanga Municipality, Ghana. Asian Journal of Poultry Science. 2015; 9(3):165–171.
- Adzitey F. Incidence and antimicrobial susceptibility of Escherichia coli isolated from beef (meat muscle, liver, and kidney) samples in Wa Abattoir, Ghana. Cog Food Agric. 2020; 6: 2–10.
- Adzitey F. Mini Review: Effect of pre-slaughter animal handling on carcass and meat quality. Int Food Res J. 2011; 18: 485-491.
- Adzitey F. Antibiotic resistance of Escherichia coli isolated from beef and its related samples in Techiman Municipality of Ghana. Asian Journal of Animal Sciences. 2015; 9: 233–240.
- Adzitey F, Huda N, Shariff AHM. Phenotypic Antimicrobial Susceptibility of Escherichia coli from Raw Meats, Ready-to-Eat Meats, and Their Related Samples in One Health Context. Microorganisms. 2021 Feb 5;9(2):326. doi: 10.3390/microorganisms9020326. PMID: 33562804; PMCID: PMC7914781.
- Hasan M, Wang J, Ahn J. Ciprofloxacin and Tetracycline Resistance Cause Collateral Sensitivity to Aminoglycosides in Salmonella Typhimurium. Antibiotics (Basel). 2023 Aug 18;12(8):1335. doi: 10.3390/antibiotics12081335. PMID: 37627755; PMCID: PMC10451331.
- Abd El-Tawab AA, Maarouf AA, Fatma I, El-Said A. Bacteriological studies on some food-borne bacteria isolated from chicken meat and meat products in Kaliobia Governorate. BVMJ. 2015; 29(2): 47-59.
- Adetunji VO, Ishola TO. Antibiotic resistance of Escherichia coli, Listeria and Salmonella isolates from retail meat tables in Ibadan municipal abattoir, Nigeria. African Journal of Biotechnology. 2011; 10(30):5795-5799.
- Adzitey F, Teye GA, Amoako DG. Prevalence, phylogenomic insights, and phenotypic characterization of Salmonella enterica isolated from meats in the Tamale metropolis of Ghana. Food Sci Nutr. 2020 May 22;8(7):3647-3655. doi: 10.1002/fsn3.1647. PMID: 32724627; PMCID: PMC7382109.
- Pławińska-Czarnak J, Wódz K, Kizerwetter-Świda M, Bogdan J, Kwieciński P, Nowak T, Strzałkowska Z, Anusz K. Multi-Drug Resistance to Salmonella spp. When Isolated from Raw Meat Products. Antibiotics (Basel). 2022 Jun 29;11(7):876. doi: 10.3390/antibiotics11070876. PMID: 35884130; PMCID: PMC9311972.
- Saud B, Paudel G, Khichaju S, Bajracharya D, Dhungana G, Awasthi MS, Shrestha V. Multidrug-Resistant Bacteria from Raw Meat of Buffalo and Chicken, Nepal. Vet Med Int. 2019 May 2;2019:7960268. doi: 10.1155/2019/7960268. PMID: 31186828; PMCID: PMC6521380.
- Martínez-Vázquez AV, Rivera-Sánchez G, Lira-Méndez K, Reyes-López MÁ, Bocanegra-García V. Prevalence, antimicrobial resistance and virulence genes of Escherichia coli isolated from retail meat in Tamaulipas, Mexico. J Glob Antimicrob Resist. 2018 Sep;14:266-272. doi: 10.1016/j.jgar.2018.02.016. Epub 2018 Mar 6. PMID: 29501529.
- Adzitey F, Assoah-Peprah P, Teye GA, Somboro AM, Kumalo HM, Amoako DG. Prevalence and Antimicrobial Resistance of Escherichia coli Isolated from Various Meat Types in the Tamale Metropolis of Ghana. Int J Food Sci. 2020 Nov 12;2020:8877196. doi: 10.1155/2020/8877196. PMID: 33274191; PMCID: PMC7676974.
- Ekli R, Adzitey F, Agbolosu AA. Farmers’ knowledge in antibiotic usage, antibiotic residues, and susceptibility of Salmonella enterica in beef samples from the Wa Municipality, Ghana. Bulletin of Animal Health and Production in Africa. 2020; 68: 89-101.
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.


 Save to Mendeley
Save to Mendeley
